Click here for Geppetto
Click here for Jiminy
Research Overview
Research in the Gormley Lab aims to use robotics plus machine learning to develop self-assembling nanomaterials that interface with and manipulate biological materials such as proteins. See the research topics below.
AI / ml guided robotics for Materials chemistry
We are programming robotics with artificial intelligence (AI) / machine learning (ML) for next-generation material chemistry.
NANOMEDICINE
This research currently focuses on designing bioactive nanomedicines for the treatment of disease.
SELF-ASSEMBLY
The Gormley Lab also investigates fundamental advancements in macromolecular self-assembly in order to better structure nanomaterials in solution.
Lab Members
Dr. Adam Gormley
Adam Gormley is an Associate Professor of Biomedical Engineering at Rutgers University and an expert in nanobiomaterials. Prior to Rutgers, Adam was a Marie Skłodowska-Curie Research Fellow at the Karolinska Institutet (2016) and a Whitaker International Scholar at Imperial College London (2012-2015) in the laboratory of Professor Molly Stevens. He obtained his PhD in Bioengineering from the University of Utah in the laboratory of Professor Hamid Ghandehari (2012), and a BS in Mechanical Engineering from Lehigh University (2006). In January 2017, Adam started the Gormley Lab which seeks to develop bioactive nanobiomaterials for therapeutic and regenerative medicine applications.
Research Faculty
Dr. Sanjeeva Murthy
Sanjeeva Murthy is a Materials Scientist working with polymers and biomaterials. He is currently focused on developing structure-property-function relationships in protein-polymer conjugates using by using light, X-ray and neutron scattering for applications such as protein-structure stabilization, drug delivery and sensors. He has a Ph.D. in Materials Science from the University of Connecticut. He did his postdoctoral work at CMU on biological cell membranes and at MIT and on artificial skin. After working for 21 years at the Corporate Research Laboratories of Honeywell, and as an Associate Professor of Physics at the University of Vermont for 4.5 years, he moved to Rutgers in 2007 as an Associate Research Professor in the department of Chemistry and Chemical Biology. At Rutgers, his research included fabrication of tissue scaffolds for use as biomedical implants, modification of surfaces to control cell-substrate interactions, and studies of degradation, mechanical behavior, biological activity and drug delivery. He has nearly 250 publications and book chapters. He is a Fellow of the American Physical Society and International Center for Diffraction Data, and a member of the American Chemical Society and Society for Biomaterials.
Post-Docs
Dr. Chris Radford
Chris Radford joined the Gormley Lab in April 2022. He received B.S. degrees in Biomedical Engineering and Economics from Duke University, and M.S. and Ph.D. degrees in Biomedical Engineering from the University of Utah. During his graduate work, he developed bioactive, cell receptor-clustering polymer conjugates and evaluated their potential as anticancer nanomedicines. In the Gormley Lab, he is interested in leveraging high-throughput polymer synthesis to further explore the role of ligand presentation in modulating the nano-bio interface to drive cellular responses.
Gantt Meredith
Gantt Meredith joined the Gormley Lab in August 2025. He received a BS in Biomedical Engineering and a BA in Linguistics from the University of Florida in 2020, and he received his PhD in Biomedical Engineering from George Washington University in 2025. In collaboration with Children’s National Hospital and the European Institute of Technology, he developed theranostic ultrasound models for the amelioration of pediatric diabetes, combining medical imaging, machine learning, and physical modeling in diagnostic clinical trials. In the Gormley Lab, he is interested in developing autonomous protein-polymer-hybrid synthesis workflows to expedite the development of pharmaceutical platforms.
Supraja Chittari
Supraja Chittari joined the Gormley Lab in September 2025. She did her both her B.S. (University of Virginia, go Hoos!) and her M.S./Ph.D. (University of North Carolina, Chapel Hill) in Chemistry. Her graduate work was at the interface of polymer and computational chemistry, developing statistical tools to quantify fundamental principles in biomimetic polymers that speak to global challenges in sustainability. In the Gormley Lab, she is interested in leveraging automated workflows to design stimulus-responsive materials for targeted therapeutic release.
PhD Students
Eugene Cheong
Eugene is a PhD student in Rutgers University studying Biomedical Engineering. He graduated from University of Massachusetts Amherst in 2020 with a B.S. in Biochemistry and Molecular Biology. Eugene’s previous research focused on using tissue engineering to create a biomimetic bone microenvironment. Over the course of his graduate work, Eugene hopes to explore and study the use of polymers in drug delivery applications.
Elena Di Mare
Elena Di Mare is a first year PhD student at Rutgers studying Biomedical Engineering. She received her BS from the University of Illinois at Urbana-Champaign with a major in Materials Science and Engineering, a focus on biomaterials, and a minor in Italian. Elena has primarily performed research on biomimetic materials. At Rutgers, she is interested in studying biological nanomaterials for drug delivery applications.
Eman Ahmed
Eman joined Rutgers Biomedical Engineering Department as a PhD student in 2022. She has a bachelor’s in biomedical engineering from Rutgers and also a minor in mathematics. She conducted research on nanoparticles characteristics in vitro and in silico. As a PhD student, she is interested in leveraging machine learning and nanobiomaterials for enhancing drug delivery.
Gabriela Tirado-Mansilla
Gabriela is a PhD student who joined the Gormley lab in 2022. She received a BS in Biomedical Engineering with a concentration in Biomechanics and Mechanobiology at the University of Connecticut in 2021. She plans on researching the relationship between the structure and chemistry of ligands to improve synthetic design of multivalent ligands. The overall goal is to design programable multivalent ligands and apply them to drug targeting, protein mimetics and more.
Weronika Wasniowska
Weronika is a PhD student studying Biomedical Engineering. She joined the Gormley Lab in 2023, after receiving her B.S. in BME from Rutgers University. While completing her undergraduate degree, she worked with electrospray deposition to optimize drug delivery using microneedle arrays. Weronika's interests lie in applying engineering methods to neurodegenerative diseases and cancers. During her PhD, she hopes to explore targeted delivery of cancer drugs utilizing photodynamic therapy.
Cristian Lopez
Cristian Lopez is a PhD student who joined the Gormley Lab in 2024. He received his B.S. in Biomedical Engineering at the University of Connecticut, where he primarily worked in developing non-viral polymeric drug delivery vehicles for CNS delivery. At the Gormley lab, his work has primarily focused on the development of self-driving labs for autonomous, high-fidelity pipelines for stabilizing monoclonal antibodies for high concentration regimes.
Masters Students
Apostolos Maroulis
Apostolos is an undergraduate studying Biomedical Engineering at Rutgers University and is currently in the Combined Degree Program for his Master of Science. His current research is focused on automating the separation and purification of polymers using Size Exclusion Chromatography through the James J. Slade Scholars program.
Dylan Waynor
Dylan is an undergraduate studying Biomedical Engineering at Rutgers University pursuing a Combined Degree program along with participating in the James J. Slade Scholars program. His current research is focused on microfluidic systems for drug encapsulation.
Automation-assisted photoinduced atom transfer radical polymerization
Oxygen-tolerant reversible-deactivation radical polymerizations (RDRP) now allow many of these reactions to proceed in open labware, such as well plates. This enables the high-throughput synthesis of tailored polymers and lowers the knowledge barrier required to obtain these materials. Building on our previous work automating photoinduced electron/energy transfer reversible addition–fragmentation chain transfer (PET–RAFT) and enzyme-assisted RAFT (Enz-RAFT) polymerization, we now introduce automated atom transfer radical polymerization (ATRP). Here, we demonstrate the potential of this platform for the high-throughput optimization of ATRP chemistry. Furthermore, we demonstrate that this workflow can help provide insights into the selection of reaction components, such as ligands and initiators, for the polymerization of kinetically difficult monomers such as methyl methacrylate with smaller rates of propagation than acrylates. This coupling paves the way for data-driven optimization of ATRP reactions, accelerated by the generation of high-throughput data sets. To facilitate the integration of robotics for high-throughput applications in polymer synthesis and optimization of photo-ATRP, we have made a Python package available to assist with experimental planning. The tool accepts Excel sheets with user-defined molar ratios, target monomer concentrations, and reagent stock concentrations and outputs an Excel sheet with actionable recipes that can be readily implemented in liquid handling transfer steps via manual or automated pipetting.
Small-angle x-ray scattering (SAXS) is a powerful technique for assessing macromolecular structure. High-throughput SAXS is limited by the time-consuming and, at times, subjective nature of SAXS data interpretation. We present SAXS Assistant, a Python-based script that streamlines SAXS data analysis to extract features for machine learning (ML) and key structural parameters, including the Guinier radius of gyration (Rg), pair distance distribution function (PDDF)-derived Rg, maximum particle dimension (Dmax), and Kratky plots. The script builds upon BioXTAS RAW and validates reliability via Guinier/PDDF Rg agreement, an important indicator of well-measured data sets. For assistance in Dmax estimation, a multilayer perceptron regressor was trained with 1940 data files from the Small Angle Scattering Biological Data Bank. The model achieved a test set performance R2 = 0.90 and mean absolute error = 11.7 Å. Training exclusively with experimental data translates analyses from researchers, including experts in the field, to the ML model, which helps assess Dmax estimations from PDDF. Gaussian mixture model clustering was implemented to classify profiles into structural classes based on entries in the Small Angle Scattering Biological Data Bank. Users may therefore assess the similarity between experimental samples and known biomolecular shapes within the mapped repository entries. This probabilistic clustering aids in quantifying information from Kratky and generating shape-descriptive features. SAXS Assistant accelerates SAXS data analysis through enforced quality control, ML-ready outputs, and flags for low-confidence results. In addition to providing the ability to analyze large data sets at high throughput, this tool is versatile and may serve researchers in both biological and synthetic polymer research fields.
C. Ramirez, et al., 2025. Biophysical Journal. 124: 3772-3786.
Data-driven design of novel polymer excipients for pharmaceutical amorphous solid dispersions
About 90% of active pharmaceutical ingredients (APIs) in the oral drug delivery system pipeline have poor aqueous solubility and low bioavailability. To address this problem, amorphous solid dispersions (ASDs) embed hydrophobic APIs within polymer excipients to prevent drug crystallization, improve solubility, and increase bioavailability. There are a limited number of commercial polymer excipients, and the structure–function relationships which lead to successful ASD formulations are not well-documented. There are, however, certain solid-state ASD characteristics that inform ASD performance. One characteristic shared by successful ASDs is a high glass transition temperature (Tg), which correlates with higher shelf stability and decreased drug crystallization. We aim to identify how polymer features such as side chain geometry, backbone methylation, and hydrophilic–lipophilic balance impact Tg to design copolymers capable of forming high-Tg ASDs. We tested a library of 50 ASD formulations (18 previously studied and 32 newly synthesized) of the model drug probucol with copolymers synthesized through automated photoinduced electron/energy transfer-reversible addition–fragmentation chain-transfer (PET-RAFT) polymerization. A machine learning (ML) algorithm was trained on the Tg data to identify the major factors influencing Tg, including backbone methylation and nonlinear side chain geometry. In both polymer alone and probucol-loaded ASDs, a Random Forest Regressor captured structure–function trends in the data set and accurately predicted Tg with an average R2 > 0.83 across a 10-fold cross validation. This ML model will be used to predict novel copolymers to design ASDs with high Tg, a crucial factor in predicting ASD success.
E.J. Di Mare, et al., 2024. Bioconjugate Chemistry. 35: 1363-1372.
Mapping biomaterial complexity by machine learning
Biomaterials often have subtle properties that ultimately drive their bespoke performance. Given this nuanced structure-function behavior, the standard scientific approach of one experiment at a time or design of experiment (DOE) methods is largely inefficient for the discovery of complex biomaterials. More recently, high-throughput experimentation coupled with machine learning methods have matured beyond expert users allowing scientists and engineers from diverse backgrounds to access these powerful data science tools. As a result, we now have the opportunity to strategically utilize all available data from high-throughput experiments to train efficacious models and map the structure-function behavior of biomaterials for their discovery. Herein, we discuss this necessary shift to data-driven determination of structure-function properties of biomaterials as we highlight how machine learning is leveraged in identifying physicochemical cues for biomaterials in tissue engineering, gene delivery, drug delivery, protein stabilization, and antifouling materials. We also discuss data-mining approaches that are coupled with machine learning to map biomaterial functions that reduce the load on experimental approaches for faster biomaterial discovery. Ultimately, harnessing the prowess of machine learning will lead to accelerated discovery and development of optimal biomaterial designs.
E. Ahmed, et al., 2024. Tissue Engineering Part A. 30: 662-680.
Machine learning in drug delivery
For decades, drug delivery scientists have been performing trial-and-error experimentation to manually sample parameter spaces and optimize release profiles through rational design. To enable this approach, scientists spend much of their career learning nuanced drug-material interactions that drive system behavior. In relatively simple systems, rational design criteria allow us to fine tune release profiles and enable efficacious therapies. However, as materials and drugs become increasingly sophisticated and their interactions have non-linear and compounding effects, the field is suffering the Curse of Dimensionality which prevents us from comprehending complex structure-function relationships. In the past, we have embraced this complexity by implementing high-throughput screens to increase the probability of finding ideal compositions. However, this brute force method was inefficient and led many to abandon these fishing expeditions. Fortunately, methods in data science including artificial intelligence / machine learning (AI/ML) are providing ideal analytical tools to model this complex data and ascertain quantitative structure-function relationships. In this Oration, I speak to the potential value of data science in drug delivery with particular focus on polymeric delivery systems. Here, I do not suggest that AI/ML will simply replace mechanistic understanding of complex systems. Rather, I propose that AI/ML should be yet another useful tool in the lab to navigate complex parameter spaces. The recent hype around AI/ML is breathtaking and potentially over inflated, but the value of these methods is poised to revolutionize how we perform science. Therefore, I encourage readers to consider adopting these skills and applying data science methods to their own problems. If done successfully, I believe we will all realize a paradigm shift in our approach to drug delivery.
A.J. Gormley, 2024. Journal of Controlled Release. 373: 23-30.
Most synthetic polypeptides are created either in the solid phase or by polymerizing cyclic amino acids. While these methods are well established, the air sensitivity of these reactions limits their implementation in high-throughput workflows despite monumental efforts. To solve this problem, Hua Lu and colleagues develop a unique and versatile approach by first synthesizing large batches of simple, reactive polypeptides. In their design, reactivity is provided by installing a selenolate group in each repeating unit of the polymer backbone. Then, each reactive group is modified using a library of electrophilic side chains. This process of post-polymerization modification is extremely efficient and enables these reactions to proceed in well plates using liquid handling robotics. The result is a simple and high-throughput process for generating highly diverse polypeptide polymers.
A Fully Automated Platform for Photoinitiated RAFT Polymerization
Oxygen tolerant polymerizations including Photoinduced Electron/Energy Transfer-Reversible Addition-Fragmentation Chain-Transfer (PET-RAFT) polymerization allow for high-throughput synthesis of diverse polymer architectures on the benchtop in parallel. Recent developments have further increased throughput using liquid handling robotics to automate reagent handling and dispensing into well plates thus enabling the combinatorial synthesis of large polymer libraries. Although liquid handling robotics can enable automated polymer reagent dispensing in well plates, photoinitiation and reaction monitoring require automation to provide a platform that enables the reliable and robust synthesis of various polymer compositions in high-throughput where polymers with desired molecular weights and low dispersity are obtained. Here, we describe the development of a robotic platform to fully automate PET-RAFT polymerizations and provide individual control of reactions performed in well plates. On our platform, reagents are automatically dispensed in well plates, photoinitiated in individual wells with a custom-designed lightbox until the polymerizations are complete, and monitored online in real-time by tracking fluorescence intensities on a fluorescence plate reader, with well plate transfers between instruments occurring via a robotic arm. We found that this platform enabled robust parallel polymer synthesis of both acrylate and acrylamide homopolymers and heteropolymers, with high monomer conversions and low dispersity. The successful polymerizations obtained on this platform make it an efficient tool for combinatorial polymer chemistry. In addition, with the inclusion of machine learning protocols to help navigate the polymer space towards specific properties of interest, this robotic platform can ultimately become a self-driving lab that can dispense, synthesize, and monitor large polymer libraries.
Polymer-protein hybrids can be deployed to improve protein solubility and stability in denaturing environments. While previous work used robotics and active machine learning to inform new designs, further biophysical information is required to ascertain structure–function behavior. Here, we show the value of tandem small-angle x-ray scattering (SAXS) and quartz crystal microbalance with dissipation (QCMD) experiments to reveal detailed polymer-protein interactions with horseradish peroxidase (HRP) as a test case. Of particular interest was the process of polymer-protein complex formation under thermal stress whereby SAXS monitors formation in solution while QCMD follows these dynamics at an interface. The radius of gyration (Rg) of the protein as measured by SAXS does not change significantly in the presence of polymer under denaturing conditions, but thickness and dissipation changes were observed in QCMD data. SAXS data with and without thermal stress were utilized to create bead models of the potential complexes and denatured enzyme, and each model fit provided insight into the degree of interactions. Additionally, QCMD data demonstrated that HRP deforms by spreading upon surface adsorption at low concentration as shown by longer adsorption times and smaller frequency shifts. In contrast, thermally stressed and highly inactive HRP had faster adsorption kinetics. The combination of SAXS and QCMD serves as a framework for biophysical characterization of interactions between proteins and polymers which could be useful in designing polymer-protein hybrids.
R. Upadhya, et al., 2022. Journal of Biomedical Materials Research Part A. 111: 440-450.
A USER’S GUIDE TO MACHINE LEARNING FOR POLYMERIC BIOMATERIALS
The development of novel biomaterials is a challenging process, complicated by a design space with high dimensionality. Requirements for performance in the complex biological environment lead to difficult a priori rational design choices and time-consuming empirical trial-and-error experimentation. Modern data science practices, especially artificial intelligence (AI)/machine learning (ML), offer the promise to help accelerate the identification and testing of next-generation biomaterials. However, it can be a daunting task for biomaterial scientists unfamiliar with modern ML techniques to begin incorporating these useful tools into their development pipeline. This Perspective lays the foundation for a basic understanding of ML while providing a step-by-step guide to new users on how to begin implementing these techniques. A tutorial Python script has been developed walking users through the application of an ML pipeline using data from a real biomaterial design challenge based on group’s research. This tutorial provides an opportunity for readers to see and experiment with ML and its syntax in Python. The Google Colab notebook can be easily accessed and copied from the following URL: www.gormleylab.com/MLcolab
T. A. Meyer, et al., 2022. ACS Polymers Au. 3: 141-157.
BIOLOGIC FORMULATION IN A SELF-DRIVING BIOMATERIALS LAB
Biologics such as monoclonal antibodies (mAbs) and RNA therapeutics have revolutionized standard of care but present stability challenges due to their fragile structure. This is particularly true considering the demanding manufacturing, storage, distribution, and administration requirements that far exceed the otherwise stable biological environment from which they are derived. Therefore, the pharmaceutical industry routinely implements a suite of experiments to optimize formulations using a standard set of excipients that are known to enhance stability. While this process has been productive, the complexity of biologic-excipient interactions prevents an efficient transition to precise and tailored formulations. Recent advances in laboratory automation, high-throughput analytics, and artificial intelligence/machine learning (AI/ML) now provide a unique opportunity to fully automate the design process and provide next-generation formulations with remarkable durability. Here, we put forth a plan to develop a biomaterials acceleration platform (BioMAP) (i.e., self-driving biomaterials lab) focused initially on biologic formulation.
M. J. Tamasi, A. J. Gormley, 2022. Cell Reports Physical Science. 3, 9, 101041.
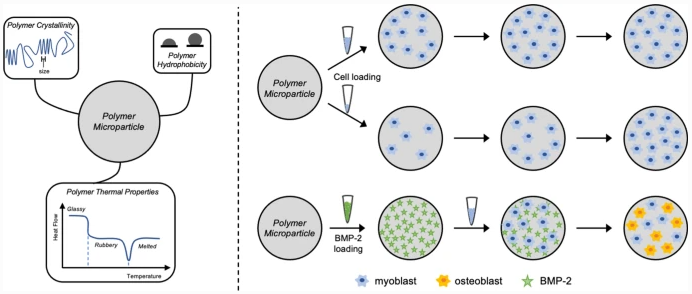
Polymer Texture Influences Cell Responses in Osteogenic Microparticles
Polymer materials used in medical devices and treatments invariably encounter cellular networks. For the device to succeed in tissue engineering applications, the polymer must promote cellular interactions through adhesion and proliferation. To predict how a polymer will behave in vitro, these material–cell interactions need to be well understood. To study polymer structure-property relationships, microparticles of four chemically distinct tyrosol-derived poly(ester-arylate) polymers and a commercially available poly(lactic acid-co-glycolic acid) (PLGA) copolymer were prepared and their interactions with cells investigated. Cell loading concentration was optimized and cell adhesion and proliferation evaluated. Particles were also tested for their ability to adsorb bone morphogenetic protein-2 (BMP-2) and differentiate a myoblast cell line towards an osteoblast lineage through BMP-2 loading and release. While cell adhesion was observed on all particles after 24 h of incubation, the highest degree of cell adhesion occurred on polymers with smaller crystallites. At longer incubation times, cells proliferated on all particle formulations, regardless of the differences in polymer properties. High BMP-2 loading was achieved for all particle formulations and all formulations showed a burst release. Even with the burst release, cells cultured on all formulations showed an upregulation in alkaline phosphatase (ALP) activity, a measure of osteoblast differentiation. As with cell adhesion, the polymer with the smaller crystallite showed the most ALP activity. We suggest that smaller crystallites serve as a proxy for topographical roughness to elicit the observed responses from cells. Furthermore, we have drawn a correlation between the polymer crystallite with the hydration potential using surface analysis techniques.
C. E. Miles, et al., 2022. Cellular and Molecular Bioengineering. 15, 409–423.
Machine Learning on a Robotic Platform for the Design of Polymer-Protein Hybrids
Polymer-protein hybrids are intriguing materials that can bolster protein stability in non-native environments, thereby enhancing their utility in diverse medicinal, commercial, and industrial applications. One stabilization strategy involves designing synthetic random copolymers with compositions attuned to the protein surface, but rational design is complicated by a vast chemical and composition space. Here, we report a strategy to design protein-stabilizing copolymers based on active machine learning, facilitated by automated material synthesis and characterization platforms. The versatility and robustness of the approach is demonstrated by the successful identification of copolymers that preserve, or even enhance, the activity of three chemically distinct enzymes following exposure to thermal denaturing conditions. Although systematic screening results in mixed success, active learning appropriately identifies unique chemistries for each enzyme. Overall, this work broadens our capabilities to design fit-for-purpose synthetic copolymers that promote or otherwise manipulate protein activity, with extensions towards the design of robust polymer-protein hybrid materials.
M. Tamasi, et al., 2022. Advanced Materials. 34, 30, 2201809.
Machine-Assisted Discovery of Chondroitinase ABC Complexes toward Sustained Neural Regeneration
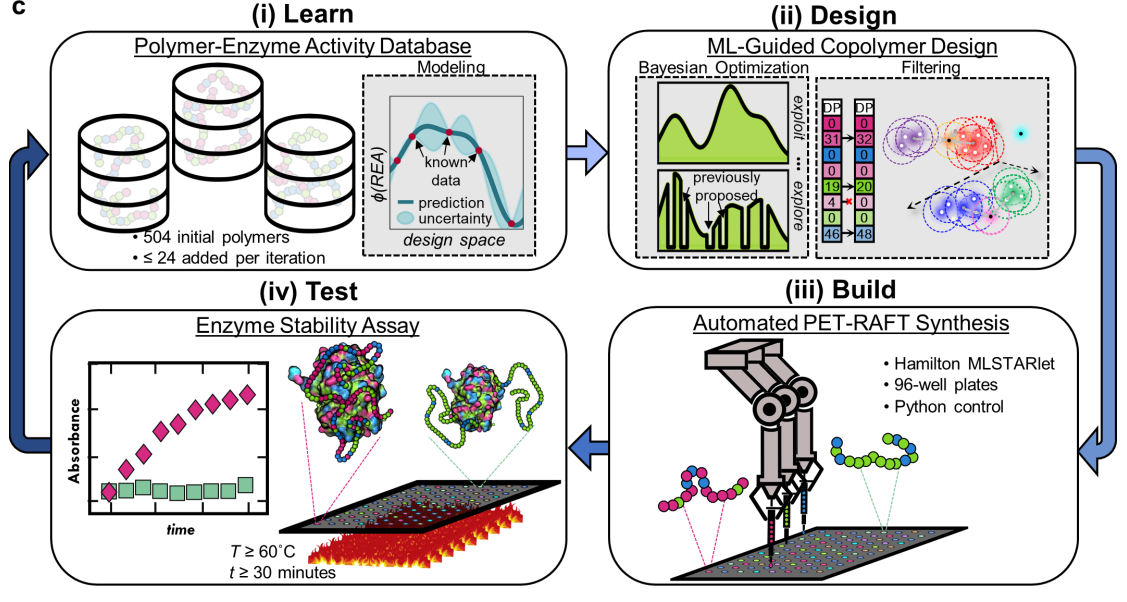
Among the many molecules that contribute to glial scarring, chondroitin sulfate proteoglycans (CSPGs) are known to be potent inhibitors of neuronal regeneration. Chondroitinase ABC (ChABC), a bacterial lyase, degrades the glycosaminoglycan (GAG) side chains of CSPGs and promotes tissue regeneration. However, ChABC is thermally unstable and loses all activity within a few hours at 37 °C under dilute conditions. To overcome this limitation, the discovery of a diverse set of tailor-made random copolymers that complex and stabilize ChABC at physiological temperature is reported. The copolymer designs, which are based on chain length and composition of the copolymers, are identified using an active machine learning paradigm, which involves iterative copolymer synthesis, testing for ChABC thermostability upon copolymer complexation, Gaussian process regression modeling, and Bayesian optimization. Copolymers are synthesized by automated PET-RAFT and thermostability of ChABC is assessed by retained enzyme activity (REA) after 24 h at 37 °C. Significant improvements in REA in three iterations of active learning are demonstrated while identifying exceptionally high-performing copolymers. Most remarkably, one designed copolymer promotes residual ChABC activity near 30%, even after one week and notably outperforms other common stabilization methods for ChABC. Together, these results highlight a promising pathway toward sustained tissue regeneration.
S. Kosuri, et al., 2022. Advanced Healthcare Materials. 11, 10, 2102101.
Control of Drug Release from Microparticles by Tuning Their Crystalline Textures: A Structure–Activity Study
Predicting drug release profiles from polymer microparticles has proven challenging due to the numerous environmental and chemical factors that affect the device and influence the rate of drug release. By measuring the various polymer properties that can influence drug release, a predictive approach can be used to select polymers with specific properties that will lead to the desired release profile for the application. To illustrate this, a library of tyrosol-derived poly(ester-arylate)s, poly(amide)s, and poly(carbonate)s were used to evaluate the effects of physical (crystallinity, water accessibility, thermal, and hydrophobicity) and chemical (polymer–drug interactions) polymer properties on the release of a highly crystalline drug dexamethasone, which was loaded at a high weight percent (wt %) in microparticles. Nuclear magnetic resonance (NMR) experiments showed that the polymer and drug were not chemically interacting and instead exist as a physical mixture even after exposure to physiological conditions. Polymer crystallinity data revealed that crystallite size was strongly correlated with faster drug release, suggesting that larger crystallites reduce the tortuosity for dexamethasone to diffuse out of the particle matrix. This correlation observed in particles with and without the drug was reproduced with bulk polymers, indicating that crystallinity data from bulk polymers can be used to predict release profiles without having to prepare drug-loaded particles. Consistent with the crystallinity data, particle pore sizes of representative formulations showed that particles with larger pores resulted in faster dexamethasone release. Interestingly, thermal properties (glass transition temperature and melting temperature), polymer hydrophobicity, and molecular weight retention at the end of the 119-day release study did not show any correlation with drug release.
C. E. Miles, et al., 2021. ACS Applied Polymer Materials. 3, 12, 6548-6561.
Self-assembly and bioconjugation in drug delivery
The delivery of drugs relies on material/biological systems that modulate the mechanism and rate of drug release. This is typically regulated by covalent and/or non-covalent interactions between the drug, the carrier material, and targeting moieties. Over the last few decades, drug delivery scientists have been leveraging the immense potential of self-assembly and bioconjugation to program these precise characteristics. In this Advanced Drug Delivery Reviews Special Theme Issue, we highlight a selection of recent examples where self-assembly and bioconjugation play a key role within this exciting and continuously evolving field.
A.J. Gormley et al., 2021. Advanced Drug Delivery Reviews. 174, 628-629
Tyrosol-Derived Biodegradable Inks with Tunable Properties for 3D Printing
Three-dimensional (3D) printing has emerged as a valuable tool in medicine over the past few decades. With a growing number of applications using this advanced processing technique, new polymer libraries with varied properties are required. Herein, we investigate tyrosol-based poly(ester-arylate)s as biodegradable inks in fused deposition modeling (FDM). Tyrosol-based polycarbonates and polyesters have proven to be useful biomaterials due to their excellent tunability, nonacidic degradation components, and the ability to be functionalized. Polymers are synthesized by polycondensation between a custom diphenol and commercially available diacids. Thermal properties, degradation rates, and mechanical properties are all tunable based on the diphenol and diacid chosen. Evaluation of material print as it relates to chemical structure, molecular weight, and thermal properties was explored. Higher-molecular-weight polymers greater than 50 kDa exhibit thermal degradation during printing and at some points are too viscous to print. It was determined that polymers with lower processing temperatures and molecular weights were printable regardless of the structure. An exception to this was pHTy6 that was printed at 65 kDa with minimal degradation. This is most likely due to its low melting temperature and, as a result, lower printing temperatures. Additionally, chemical improvements were made to incorporate thiol–alkene click chemistry as a means for postprint curing. Low-molecular-weight pHTy6 was end-capped with alkene functionality. This material was then formulated with either a dithiol for chain extension or tetrathiol for cross-linking. Scaffolds were cured after printing for 5, 15, 30 and 60 min intervals where longer cure times resulted in a tougher material. This design builds on the library of biologically active materials previously explored and aims to bring new biomaterials to the field of 3D-printed personal medicine.
J. Cohen, et al., 2021. ACS Biomaterials Science & Engineering. 7, 9, 4454-4462.
Tyrosol Derived Poly(ester-arylate)s for Sustained Drug Delivery from Microparticles
New biodegradable polymers are needed for use in drug delivery systems to overcome the high burst release, lack of sustained drug release, and acidic degradation products frequently observed in current formulations. Commercially available poly(lactide-co-glycolide) (PLGA) is often used for particle drug release formulations; however, it is often limited by its large burst release and acidic degradation products. Therefore, a biocompatible and biodegradable tyrosol-derived poly(ester-arylate) library has been used to prepare a microparticle drug delivery system which shows sustained delivery of hydrophobic drugs. Studies were performed using polymers with varying hydrophilicity and thermal properties and compared to PLGA. Various drug solubilizing cosolvents were used to load model drugs curcumin, dexamethasone, nicotinamide, and acyclovir. Hydrophobic drugs curcumin and dexamethasone were successfully loaded up to 50 weight percent (wt %), and a linear correlation between drug wt % loaded and the particle glass transition temperature (Tg) was observed. Both curcumin and dexamethasone were visible on the particle surface at 20 wt % loading and higher. By adjusting the polymer concentration during particle formation, release rates were able to be controlled. Release studies of dexamethasone loaded particles with a lower polymer concentration showed a biphasic release profile and complete release after 47 days. Particles prepared using a higher polymer concentration showed sustained release for up to 77 days. Comparably, PLGA showed a traditional triphasic release profile and complete release after 63 days. This novel tyrosol-derived poly(ester-arylate) library can be used to develop injectable, long-term release formulations capable of providing sustained drug delivery.
C.E. Miles, et al., 2021. ACS Biomaterials Science & Engineering. 7, 2580-2591.
machine learning in combinatorial polymer chemistry
The design of new functional polymers depends on the successful navigation of their structure-function landscapes. Advances in combinatorial polymer chemistry and machine learning provide exciting opportunities for the engineering of fit-for-purpose polymeric materials.
A.J. Gormley and M.A. Webb, 2021. Nature Reviews Materials. 6, 642-644.
Automated PET-RAFT Polymerization toward Pharmaceutical Amorphous Solid Dispersion Development
In pharmaceutical oral drug delivery development, about 90% of drugs in the pipeline have poor aqueous solubility leading to severe challenges with oral bioavailability and translation to effective and safe drug products. Amorphous solid dispersions (ASDs) have been utilized to enhance the oral bioavailability of poorly soluble active pharmaceutical ingredients (APIs). However, a limited selection of regulatory-approved polymer excipients exists for the development and further understanding of tailor-made ASDs. Thus, a significant need exists to better understand how polymers can be designed to interact with specific API moieties. Here, we demonstrate how an automated combinatorial library approach can be applied to the synthesis and screening of polymer excipients for the model drug probucol. We synthesized a library of 25 random heteropolymers containing one hydrophilic monomer (2-hydroxypropyl acrylate (HPA)) and four hydrophobic monomers at varied incorporation. The performance of ASDs made by a rapid film casting method was evaluated by dissolution using ultra-performance liquid chromatography (UPLC) sampling at various time points. This combinatorial library and rapid screening strategy enabled us to identify a relationship between polymer hydrophobicity, monomer hydrophobic side group geometry, and API dissolution performance. Remarkably, the most effective synthesized polymers displayed slower drug release kinetics compared to industry standard polymer excipients, showing the ability to modulate the drug release profile. Future coupling of high throughput polymer synthesis, high throughput screening (HTS), and quantitative modeling would enable specification of designer polymer excipients for specific API functionalities.
R. Upadhya, et al., 2021. ACS Applied Polymer Materials. 3, 3, 1525-1536
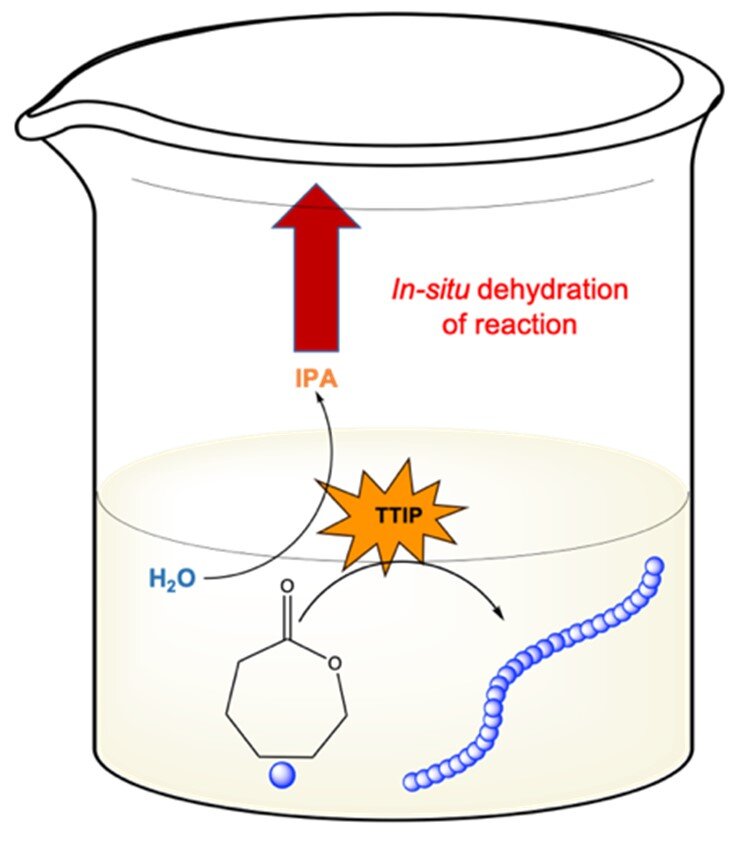
Ring Opening Polymerization of ɛ-Caprolactone Through Water
Ring opening polymerization (ROP) is commonly used to synthesize biodegradable polymers such as polycaprolactone (PCL). These reactions typically demand anhydrous reagents and inert atmosphere, only attainable through the of including glove boxes and Schlenk lines. Because of these practical limitations, ROP is typically inaccessible to the non-expert. Herein, we have developed two techniques for limited ROP of ɛ-caprolactone (CL) without previous dryin`g of reagents by simply conducting the reaction in a laboratory oven. In the first method, a vacuum oven was used to evaporate water from a traditional ROP reaction with stannous octoate (Sn(Oct)2) as the catalyst. In the second method, we ‘polymerize through’ water using titanium isopropoxide (TTIP) to simultaneously quench residual water and catalyze ROP. Using these two methods, we achieved variable chain length (degree of polymerization (DP) 25-500) and molecular weight distribution (Đ = 1.3-1.5) of PCL. We found that ROP with TTIP works better for higher molecular weights up to DP 500 than Sn(Oct)2 under vacuum. Moreover, we investigated the living nature of ROP with TTIP where a block copolymer of PCL-PLA was achieved.
Automation and data-driven design of polymer therapeutics.
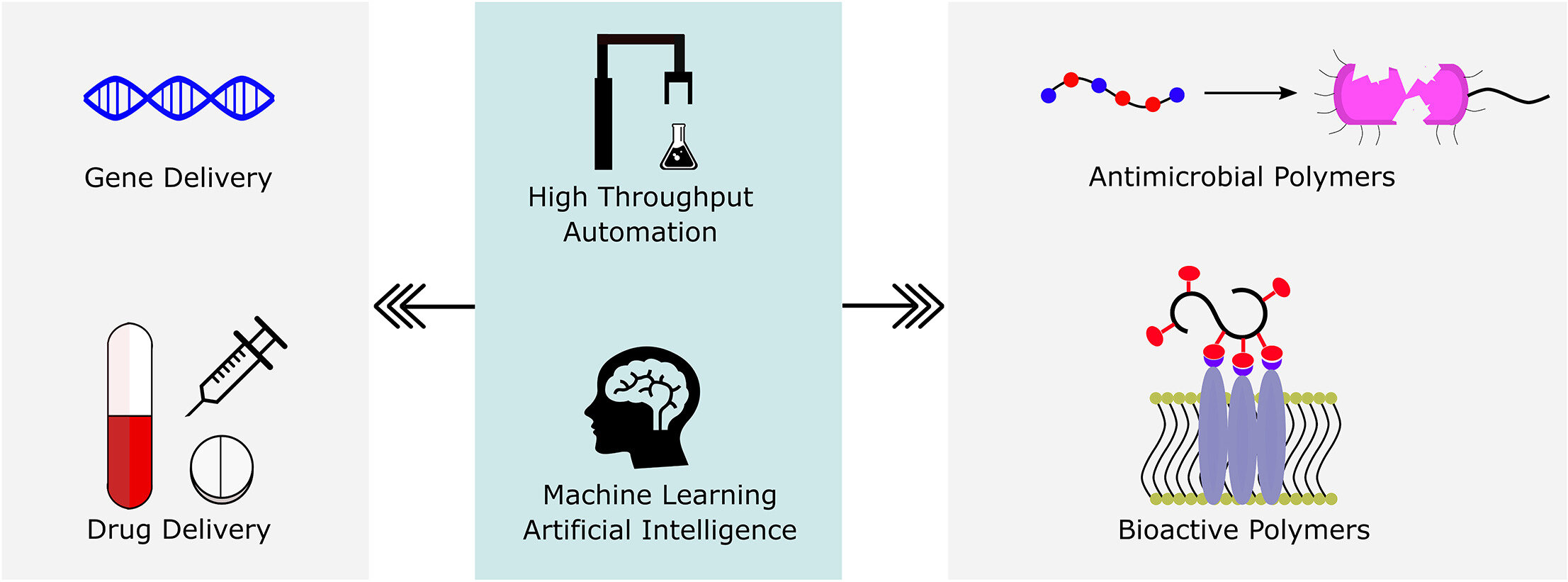
Polymers are uniquely suited for drug delivery and biomaterials applications due to tunable structural parameters such as length, composition, architecture, and valency. To facilitate designs, researchers may explore combinatorial libraries in a high throughput fashion to correlate structure to function. However, traditional polymerization reactions including controlled living radical polymerization (CLRP) and ring-opening polymerization (ROP) require inert reaction conditions and extensive expertise to implement. With the advent of air-tolerance and automation, several polymerization techniques are now compatible with well plates and can be carried out at the benchtop, making high throughput synthesis and high throughput screening (HTS) possible. To avoid HTS pitfalls often described as “fishing expeditions,” it is crucial to employ intelligent and big data approaches to maximize experimental efficiency. This is where the disruptive technologies of machine learning (ML) and artificial intelligence (AI) will likely play a role. In fact, ML and AI are already impacting small molecule drug discovery and showing signs of emerging in drug delivery. In this review, we present state-of-the-art research in drug delivery, gene delivery, antimicrobial polymers, and bioactive polymers alongside data-driven developments in drug design and organic synthesis. From this insight, important lessons are revealed for the polymer therapeutics community including the value of a closed loop design-build-test-learn workflow. This is an exciting time as researchers will gain the ability to fully explore the polymer structural landscape and establish quantitative structure-property relationships (QSPRs) with biological significance.
R. Upadhya, et al., 2021. Advanced Drug Delivery Reviews. 171, 1-28.
Automation of Controlled/Living Radical Polymerization
Controlled living/radical polymerization (CLRP) techniques are widely utilized to synthesize advanced and controlled synthetic polymers for chemical and biological applications. While automation has long stood as a high throughput (HTP) research tool to increase productivity as well as synthetic/analytical reliability and precision, oxygen intolerance of CLRP has limited the widespread adoption of these systems. Recently, however, oxygen tolerant CLRP techniques such as oxygen tolerant PET‐RAFT, Enz‐RAFT, and ATRP have emerged. Here, we demonstrate the use of a Hamilton MLSTARlet liquid handling robot for automating CLRP reactions. Synthesis processes were developed using Python and used to automate reagent handling, dispensing sequences, and synthesis steps required to create homopolymers, random heteropolymers, and block copolymers in 96 well plates, as well as post‐polymerization modifications. Using this approach, we demonstrate the synergy between highly customizable liquid handling robotics and oxygen tolerant CLRP to automate advanced polymer synthesis for HTP and combinatorial polymer research.
M. Tamasi, et al., 2020. Advanced Intelligent Systems. 2, 1900126.
A Dual Wavelength Polymerization and Bioconjugation Strategy for High Throughput Synthesis of Multivalent Ligands
Structure–function relationships for multivalent polymer scaffolds are highly complex due to the wide diversity of architectures offered by such macromolecules. Evaluation of this landscape has traditionally been accomplished case-by-case due to the experimental difficulty associated with making these complex conjugates. Here, we introduce a simple dual-wavelength, two-step polymerize and click approach for making combinatorial conjugate libraries. It proceeds by incorporation of a polymerization friendly cyclopropenone-masked dibenzocyclooctyne into the side chain of linear polymers or the α-chain end of star polymers. Polymerizations are performed under visible light using an oxygen tolerant porphyrin-catalyzed photoinduced electron/energy transfer-reversible addition–fragmentation chain-transfer (PET-RAFT) process, after which the deprotection and click reaction is triggered by UV light. Using this approach, we are able to precisely control the valency and position of ligands on a polymer scaffold in a manner conducive to high throughput synthesis.
Z. Li*, S. Kosuri*, et al., 2019. Journal of the American Chemical Society. 141, 50, 19823- 19830.
Purifying low-volume combinatorial polymer libraries with gel filtration columns
Recent advances in oxygen tolerant controlled/living radical polymer chemistry now enable efficient synthesis of diverse and combinatorial polymer libraries. While library synthesis has been dramatically simplified, equally efficient purification strategies for removal of small molecule impurities are not yet established in high throughput settings. It is shown that gel filtration columns for chromatography (GFC) frequently used in the protein science community are well suited for high throughput polymer purification. Using either single-use columns or gel filtration plates, we demonstrate the purification of 32 diverse polymers in a library with >95% removal of small molecule impurities and >85% polymer retention in a single purification step. Doing so replaces the typical procedure of polymer precipitation which requires solvent optimization for each polymer in a complex library. Overall, this work raises awareness in the polymer science community that gel filtration is amenable to purification of large polymer libraries and can speed up the progress of combinatorial polymer chemistry.
R. Upadhya, et al., 2019. Macromolecular Rapid Communications. 1900528.
PET-RAFT and SAXS: High Throughput Tools To Study Compactness and Flexibility of Single-Chain Polymer Nanoparticles
From protein science, it is well understood that ordered folding and 3D structure mainly arise from balanced and noncovalent polar and nonpolar interactions, such as hydrogen bonding. Similarly, it is understood that single-chain polymer nanoparticles (SCNPs) will also compact and become more rigid with greater hydrophobicity and intrachain hydrogen bonding. Here, we couple high throughput photoinduced electron/energy transfer reversible addition–fragmentation chain-transfer (PET-RAFT) polymerization with high throughput small-angle X-ray scattering (SAXS) to characterize a large combinatorial library (>450) of several homopolymers, random heteropolymers, block copolymers, PEG-conjugated polymers, and other polymer-functionalized polymers. Coupling these two high throughput tools enables us to study the major influence(s) for compactness and flexibility in higher breadth than ever before possible. Future implementation of this combinatorial and high throughput approach for characterizing SCNPs will allow for the creation of detailed design parameters for well-defined macromolecular chemistry.
Living in the Fast Lane—High Throughput Controlled/Living Radical Polymerization
Combinatorial and high throughput (HTP) methodologies have long been used by the pharmaceutical industry to accelerate the rate of drug discovery. HTP techniques can also be applied in polymer chemistry to more efficiently elucidate structure–property relationships, to increase the speed of new material development, and to rapidly optimize polymerization conditions. Controlled living/radical polymerization (CLRP) is widely employed in the preparation of potential materials for bioapplications being suitable for a large variety of polymeric materials with various architectures. The versatility of CLRP makes it an ideal candidate for combinatorial and HTP approaches to research, and recently, the development of oxygen tolerant CLRP techniques has greatly simplified the methodology. In this Perspective, we provide an overview of conventional CLRP, including automated parallel synthesizers, as well as oxygen tolerant CLRP applications for HTP polymer research.
UP IN THE AIR: TOLERANCE TO OXYGEN IN CONTROLLED/LIVING RADICAL POLYMERISATION
The requirement for deoxygenation in controlled/living radical polymerisation (CLRP) places significant limitations on its widespread implementation by necessitating the use of large reaction volumes, sealed reaction vessels as well as requiring access to specialised equipment such as a glove box and/or inert gas source. As a result, in recent years there has been intense interest in developing strategies for overcoming the effects of oxygen inhibition in CLRP and therefore remove the necessity for deoxygenation. In this review, we highlight several strategies that have already been proposed for achieving oxygen tolerant CLRP including: “polymerising through” oxygen, enzyme mediated deoxygenation and the continuous regeneration of a redox-active catalyst. In order to provide further clarity to the field, we also establish some basic parameters for evaluating the degree of “oxygen tolerance” that can be achieved using a given oxygen scrubbing strategy. Finally, we propose some applications that could most benefit from the implementation of oxygen tolerant CLRP and provide a perspective on the future direction of this field.
J. Yeow, et al., 2018. Chemical Society Reviews. 47: 4357-4387.
An oxygen-Tolerant PET-RAFT Polymerization for screening structure-Activity Relationships
The complexity of polymer–protein interactions makes rational design of the best polymer architecture for any given biointerface extremely challenging, and the high throughput synthesis and screening of polymers has emerged as an attractive alternative. A porphyrin-catalysed photoinduced electron/energy transfer–reversible addition-fragmentation chain-transfer (PET-RAFT) polymerisation was adapted to enable high throughput synthesis of complex polymer architectures in dimethyl sulfoxide (DMSO) on low-volume well plates in the presence of air. The polymerisation system shows remarkable oxygen tolerance, and excellent control of functional 3- and 4-arm star polymers.We then apply this method to investigate the effect of polymer structure on protein binding, in this case to the lectin concanavalin A (ConA). Such an approach could be applied to screen the structure–activity relationships for any number of polymer–protein interactions.
A.J. Gormley, et al., 2018. Angewandte Chemie Int. Ed. 57: 1-7.
COMBINATORIAL LOW-VOLUME SYNTHESIS OF WELL-DEFINED POLYMERS BY ENZYME DEGASSING
The synthesis of well-defined polymers in a low-volume, combinatorial fashion has long been a goal in polymer chemistry. Here, we report the preparation of a wide range of highly controlled homo and block co-polymers by Enz-RAFT (enzyme-assisted reversible addition–fragmentation chain transfer) polymerization in microtiter plates in the open atmosphere. The addition of 1 μmglucose oxidase (GOx) to water/solvent mixtures enables polymerization reactions to proceed in extremely low volumes (40 μL) and low radical concentrations. This procedure provides excellent control and high conversions across a range of monomer families and molecular weights, thus avoiding the need to purify for screening applications. This simple technique enables combinatorial polymer synthesis in microtiter plates on the benchtop without the need of highly specialized synthesizers and at much lower volumes than is currently possible by any other technique.
R. Chapman and A.J. Gormley, et al., 2016. Angewandte Chemie. 128: 4576-4579.
Layer-by-Layer Self-Assembly of Polymer Films and Capsules through Coiled-Coil Peptides
The layer-by-layer (LbL) technique is a simple and robust process for fabricating functional multilayer thin films. Here, we report the use of de novo designed polypeptides that self-assemble into coiled-coil structures (four-helix bundles) as a driving force for specific multilayer assembly. These pH- (sensitive between pH 4 and 7) and enzyme-responsive polypeptides were conjugated to polymers, and the LbL assembly of the polymer–peptide conjugates allowed the deposition of up to four polymer–peptide layers on planar surfaces and colloidal substrates. Stable hollow capsules were obtained, and by taking advantage of the peptide’s susceptibility to specific enzymatic cleavage, release of encapsulated cargo within the carriers can be triggered within 2 h in the presence of matrix metalloproteinase-7. The enormous diversity of materials that can form highly controllable and programmable coiled-coil interactions creates new opportunities and allows further exploration of the multilayer assembly and the formation of carrier capsules with unique functional properties.
A.J. Gormley, et al., 2015. Chemistry of Materials. 27: 5820–5824.
Polymerization Amplified Detection for Nanoparticle-Based Biosensing
Efficient signal amplification processes are key to the design of sensitive assays for biomolecule detection. Here, we describe a new assay platform that takes advantage of both polymerization reactions and the aggregation of nanoparticles to amplify signal. In our design, a cascade is set up in which radicals generated by either enzymes or metal ions are polymerized to form polymers that can entangle multiple gold nanoparticles (AuNPs) into aggregates, resulting in a visible color change. Less than 0.05% monomer-to-polymer conversion is required to initiate aggregation, providing high sensitivity toward the radical generating species. Good sensitivity of this assay toward horseradish peroxidase, catalase, and parts per billion concentrations of iron and copper is shown. Incorporation of the oxygen-consuming enzyme glucose oxidase (GOx), enables this assay to be performed in open air conditions at ambient temperature. We anticipate that such a design will provide a useful platform for sensitive detection of a broad range of biomolecules through polymerization-based amplification.
A.J. Gormley* and R. Chapman*, et al., 2014. Nano Letters. 14: 6368–6373.
Highly Controlled Open Vessel RAFT Polymerizations by Enzyme Degassing
Intolerance to oxygen is a key limitation in many reactions and particularly in controlled radical polymerizations. Here we introduce the use of enzymes such as glucose oxidase (GOx) to deoxygenate reversible addition–fragmentation chain transfer polymerizations (Enz-RAFT), facilitating the preparation of highly controlled polymers in vessels open to ambient oxygen. Because the removal of oxygen is so efficient, very low concentrations of GOx and initiator can be used, enabling excellent control which is demonstrated by pseudoliving polymerization kinetics and the preparation of multiblock copolymers with narrow molecular weight distributions (Mw/Mn < 1.15). GOx retains sufficient activity to facilitate polymerization not only in aqueous solutions but also in a range of water/organic solvent mixtures, and we demonstrate the use of this technique to perform open vessel Enz-RAFT polymerizations in various methanol and dioxane/water mixtures.
R. Chapman* and A.J. Gormley*, et al., 2014. Macromolecules. 47: 8541-8547.
K.Z. Meidell, R. Robinson, A. Vieira‐de‐Abreu, A.J. Gormley, H. Ghandehari, D.W. Grainger, R.A. Campbell, 2017. RGDfK‐functionalized gold nanorods bind only to activated platelets. Journal of Biomedical Materials Research Part A. 105: 209-217.
P. Campagnolo, A.J. Gormley, L.W. Chow, A.G. Guex, P.A. Parmar, J.L. Puetzer, J.A.M. Steele, A. Breant, P. Madeddu, M.M. Stevens, 2016. Pericyte seeded dual peptide scaffold with improved endothelialization for vascular graft tissue engineering. Advanced Healthcare Materials. 5: 3046-3055.
R.H. Harrison, J.A.M. Steele, R. Chapman, A.J. Gormley, L.W. Chow, M.M. Mahat, L. Podhorska, R.G. Palgrave, D.J. Payne, S.P. Hettiaratchy, I.E. Dunlop, M.M. Stevens, 2015. Modular and Versatile Spatial Functionalization of Tissue Engineering Scaffolds through Fiber-Initiated Controlled Radical Polymerization. Advanced Functional Materials. 25: 5748-5757.
B. Buckway, N. Frazier, A.J. Gormley, A. Ray, H. Ghandehari, 2014. Gold nanorod-mediated hyperthermia enhances the efficacy of HPMA copolymer – 90Y conjugates in treatment of prostate tumors. Nuclear Medicine and Biology. 41:282-289.
C-M. Horejs, A. Serio, A. Purvis, A.J. Gormley, S. Bertazzo, A. Poliniewicz, A.J. Wang, P. DiMaggio, E. Hohenester, M.M. Stevens, 2014. Biologically active laminin-111 fragment that modulates the epithelial-to-mesenchymal transition in embryonic stem cells. Proceedings of the National Academy of Sciences. 111: 5908-5913.
N. Larson, A.J. Gormley, N. Frazier, H. Ghandehari, 2013. Synergistic enhancement of cancer therapy using a combination of heat shock protein targeted HPMA copolymer-drug conjugates and gold nanorod induced hyperthermia. Journal of Controlled Release. 170: 41-50.
A.J. Gormley, N. Larson, A. Banisadr, R. Robinson, N. Frazier, A. Ray, H. Ghandehari, 2013. Plasmonic photothermal therapy increases the tumor mass penetration of HPMA copolymers. Journal of Controlled Release. 166: 130-138.
A.J. Gormley*, N. Larson*, A. Ray, R. Robinson, S. Sadekar, H. Ghandehari, 2012. Guided delivery of polymer therapeutics using plasmonic photothermal therapy. Nano Today. 7: 158-167.
A.J. Gormley, A. Malugin, A. Ray, R. Robinson, H. Ghandehari, 2011. Biological evaluation of RGDfK – gold nanorod conjugates for prostate cancer treatment. Journal of Drug Targeting. 19: 915-924.
A.J. Gormley, K. Greish, A. Ray, R. Robinson, J.A. Gustafson, H. Ghandehari, 2011. Gold nanorod mediated plasmonic photothermal therapy: A tool to enhance macromolecular delivery. International Journal of Pharmaceutics. 415: 315-8.
News
May 24th: In honor of Dr. Gormley’s recent tenure & Presidential Fellowship award, the lab got together to present him with a gift to commemorate the last 6 years of the Gormley Lab:
April 21st: A Massive congratulations to Dr. Gormley, who has been granted tenure at Rutgers University, and will be promoted to the title of Associate Professor in July! Additionally, Dr. Gormley was presented with The Presidential Fellowship for Teaching Excellence at Rutgers for his contributions to teaching & research during his pre-tenure years!
January 5th: Jules Lee’s final M.S. paper, “A Fully Automated Platform for Photoinitiated RAFT Polymerization”, has been published in Digital Discovery!
January 1st: Happy New Year! Here’s to 2023 being just as exciting and productive as 2022 was.
December 20th: Congratulations to Rahul, with his final Ph.D paper, “Examining Polymer-Protein Biophysical Interactions With Small-Angle X-Ray Scattering and Quartz Crystal Microbalance With Dissipation” having been published in the Journal of Biomedical Materials Research Part A!
November 17th: Congratulations to Travis, whose parting paper & Machine Learning tutorial, “A User’s Guide to Machine Learning for Polymeric Biomaterials” has been published in ACS Polymers Au. The associated Google CoLab tutorial is also available here on the Gormley lab website: http://www.gormleylab.com/MLcolab
September 21st: Matt & Dr. Gormley have compiled an excellent paper on the future perspective of self-driving biomaterials labs, which has been published in Cell Reports Physical Science.
July 27th: Not only was Matt’s paper, “Machine Learning on a Robotic Platform for the Design of Polymer–Protein Hybrids” accepted for publication by Advanced Materials, it’s now made the front page!
July 18th: Catherine Mile’s paper, “Polymer Texture Influences Cell Responses in Osteogenic Microparticles”, has been accepted & published by Cellular & Molecular Bioengineering!
May 22nd - 26th: The Gormley Lab gets around (the world)! Adam, Matthew, & Jules traveled abroad to attend the Polymer Therapeutics 2022 conference in Valencia, Spain!
May 20th, 2022: Matt’s paper, “Machine Learning on a Robotic Platform for the Design of Polymer–Protein Hybrids”, has moved on from ChemRxiv and been published in Advanced Materials!
May 18th, 2022: Shashank’s paper about ChABC made the front page of Advanced Healthcare Materials, congratulations!
April 23, 2022: The Gormley Lab went on a retreat together to Frost Valley, NY, camping & hiking in the Catskills mountains!
April 1, 2022: Welcome to our latest Postdoc, Chris Radford!
March 7, 2022: The Gormley Lab is recognized in Rutgers news for our work with ChABC: “Rutgers Researchers Harness AI and Robotics to Treat Spinal Cord Injuries” https://research.rutgers.edu/news/rutgers-researchers-harness-ai-and-robotics-treat-spinal-cord-injuries
February 28, 2022: Welcome to a new Postdoc in our lab, Prajakatta Mulay!
February 2, 2022: Congratulations to Shashank & team for the publication of your paper, “Machine-Assisted Discovery of Chondroitinase ABC Complexes Towards Sustained Neural Regeneration” in Advanced Healthcare Materials! https://doi.org/10.1002/adhm.202102101
January 24, 2022: Welcome to the lab, Laboratory Technician Jordan Eckhoff!
January 24, 2022: Congratulations to Matthew & team, your paper about the PolyTron, “Machine Learning on a Robotic Platform for the Design of Polymer-Protein Hybrids” has been published in Materials Science! 10.26434/chemrxiv-2022-x2qdz
December 14, 2021: Congratulations Heloise! Your Master's thesis, “The Stabilization of Chondroitinase ABC Through the Use of Heteropolymers for the Treatment of Spinal Cord Injuries” was approved by your defense committee!
November 19, 2021: Congratulations to Catherine! Your micropaticle drug release paper, “Control of Drug Release from Microparticles by Tuning Their Crystalline Textures: A Structure–Activity Study” was published in ACS Applied Polymer Materials! https://doi.org/10.1021/acsapm.1c01254
August 31, 2021: The Gormley Lab and colleagues here at Rutgers and Princeton have been granted a $1.8M NSF award through the Designing Materials to Revolutionize and Engineer our Future (DMREF) program! This collaborative project will use machine learning and robotics for the data-driven design of protein-polymer hybrid materials.
June 15, 2021: Welcome to the lab PhD students Elena Di Mare and Cesar Ramirez!
May 19, 2021: Congratulations Catherine on your awesome and super thorough microparticles paper was just published today in ACS Biomaterials Science & Engineering, a premier biomaterials journal!
February 15, 2021: Congratulations Rahul for publishing in ACS Applied Polymer Materials!
December 1, 2020: Congratulation Supriya on the acceptance of your paper to Polymer Chemistry! It has been selected as a HOT article by the journal, highlighting the impact of these great results!
November 12, 2020: Congratulations Rahul and team for acceptance of your review to Advanced Drug Delivery Reviews on ‘Automation and Data-Driven Design of Polymer Therapeutics’!
August 10, 2020: Huge news, the Gormley Lab has received its first NIH grant! Worth nearly $2M, we have been funded to study the effect of multivalency on optimized ligand presentation.
August 5, 2020: Our team of entrepreneurs named Plexymer have been selected to participate in the NSF I-Corps program to complete a customer discovery process. We will be participating in summer 2021.
August 1, 2020: Welcome to the lab PhD student Eugene Cheong!
June 18, 2020: The Gormley Lab has received an NSF grant from the CBET program! Title of the project is “Semi-automated discovery of synthetic polymers with protein features”. This is based on work from Rahul’s Macromolecules paper!
December 6, 2019: Fun press about our recently published robotic technology!
https://www.advancedsciencenews.com/automating-the-production-of-advanced-materials/
December 4, 2019: Two new and exciting research articles by PhD students Shashank Kosuri and Matthew Tamasi published in JACS and Advanced Intelligent Systems, respectively! Incredible accomplishments by two incredible researchers. Their next work will be even more exciting!
JACS: https://pubs.acs.org/doi/abs/10.1021/jacs.9b09899
Advanced Intelligent Systems: https://onlinelibrary.wiley.com/doi/abs/10.1002/aisy.201900126
November 18, 2019: Congratulations Rahul and Mythili on your paper published in Macromolecular Rapid Communications! A very practical report showing how to purify low-volume polymer libraries with disposable gel filtration columns.
https://onlinelibrary.wiley.com/doi/full/10.1002/marc.201900528
October 25, 2019: Congratulations Rahul on your paper being published in Macromolecules! Such an amazing piece of work that took an incredible amount of effort and dedication. A true masterpiece! https://pubs.acs.org/doi/10.1021/acs.macromol.9b01923
June 12, 2019: Excited to receive the A. Walter Tyson Assistant Professorship Award given each year to outstanding junior faculty in the School of Engineering!
January 16, 2019: Welcome to the lab Supriya Atta as the lab’s first post doc!! He joins us from Laura Fabris’ lab in Materials Science. We look forward to working with you and doing great science!
December 27, 2018: New review article from the Gormley Lab and collaborators on the front cover of Macromolecules!
September 18, 2018: Gormley Lab granted two awards from the New Jersey Health Foundation
Assistant Professor Adam Gormley has been awarded two grants from the New Jersey Health Foundation to support his lab’s work in diagnostics and polymer chemistry. The first is a $35,000 Research Grants award to support their development of low cost and point-of-care diagnostics to differentiate bacterial vs. viral infections. The second award is a $50,000 Innovation Grant to support their highly innovative and automated polymer chemistry.
August 9, 2018: The Gormley Lab had its first lab retreat hiking through the Delaware Water Gap!
July 11, 2018: Major congratulations to Matt and Rahul this week for earning prestigious fellowships!! Rahul was selected for a Biotechnology Training Program Fellowship and Matt was selected for a GAANN Fellowship! Both of these are a testament to each of their strengths as scientists and students. Very well deserved indeed!
June 21, 2018: The Gormley Lab has published an exciting review article on oxygen tolerant polymerizations in Chemical Society Reviews in collaboration with teams at the University of New South Wales. Click on the cover art to the right to access the review!
Exciting press release information on our MGH Prize for Primary Care!
http://www.infectioncontroltoday.com/diagnostics/could-paper-device-diagnose-infectious-disease
https://www.rdmag.com/news/2018/03/could-paper-device-diagnose-infectious-disease
December 20, 2017: Major congratulations to the all-star senior design team (Neel, Charles, Sarah and Sudeepti) for winning Second Place in the final MGH Student Technology Prize for Primary Care Competition with a $50,000 award!!!
October 9, 2017: Tim Mahon has joined the lab to do his Master's degree. Welcome to the lab Tim!
September 22, 2017: The Gormley Lab senior design team including Neel, Charles, Sarah and Sudeepti presented their project to the Namibian delegation including Prime Minister Saara Kuugongelwa! Such a cool opportunity to meet with a world leader and talk about their exciting work.
August 2, 2017: ACS-IRG Early Investigator Pilot Award granted to Prof. Adam Gormley
More exciting news for the Gormley Lab which has been awarded an American Cancer Society – Institutional Research Grant Early Investigator Pilot Award. This one-year $50,000 grant is helping Prof. Adam Gormley’s lab develop bioactive nanomedicines that target receptor proteins on cancer cells.
July 31, 2017: Gormley lab receives a Busch Biomedical Grant Award
The Gormley Lab has been awarded a two-year, $40,000 Busch Biomedical Grant. Dr. Gormley’s lab is developing novel polymers that can interface with and manipulate receptor proteins as therapeutics. This Busch Biomedical Award will help the Gormley Lab establish this technology and create a new generation of therapeutics using state-of-the-art materials science techniques.
July 12, 2017: The Gormley Lab had its first BBQ! It has been a great summer with Shashank, Matt, Jason, Kelsey, Phil and Rahul all pitching in!
May 30, 2017: Congratulations to the Gormly Lab senior design team for placing top 5 in the MGH Student Technology Prize for Primary Healthcare competition! Well done Neel, Charles, Sarah and Sudeepti!
Mar 2, 2017: Our newest lab member, Jason DiStefano, has joined the lab. Welcome Jason!
Jan 20, 2017: Three new students join the Gormley Lab. Welcome Elisheva, Shashank and Matt!
Jan 18, 2017: First class on Biointerfacial Characterization
Jan 1, 2017: Gormley Lab opens at Rutgers University!
contact
Gormley Lab
Biomedical Engineering
Rutgers University
BME Room 220
559 Taylor Road
Piscataway, New Jersey 08854
Adam.Gormley@rutgers.edu
Tel: +1-848-445-6569
Fax: +1-732-445-3753